About
Meet me - a passionate and driven bioinformatician with a love for exploring the secrets of life through data. With over a decade of experience in the field, I've delved into multiple projects ranging from drug target discovery to heart regeneration. My expertise lies in single-cell RNAseq analysis, trasncriptomics, lipidomics, machine learning, network analysis, and multiomics data integration. I've developed and hosted multiple shiny apps for visualization and accessibility by non-programmers!
But my skills don't just stop there - I'm also proficient in pipeline development, motif finding, topic modeling, natural language processing, and even spatial transcriptomics!
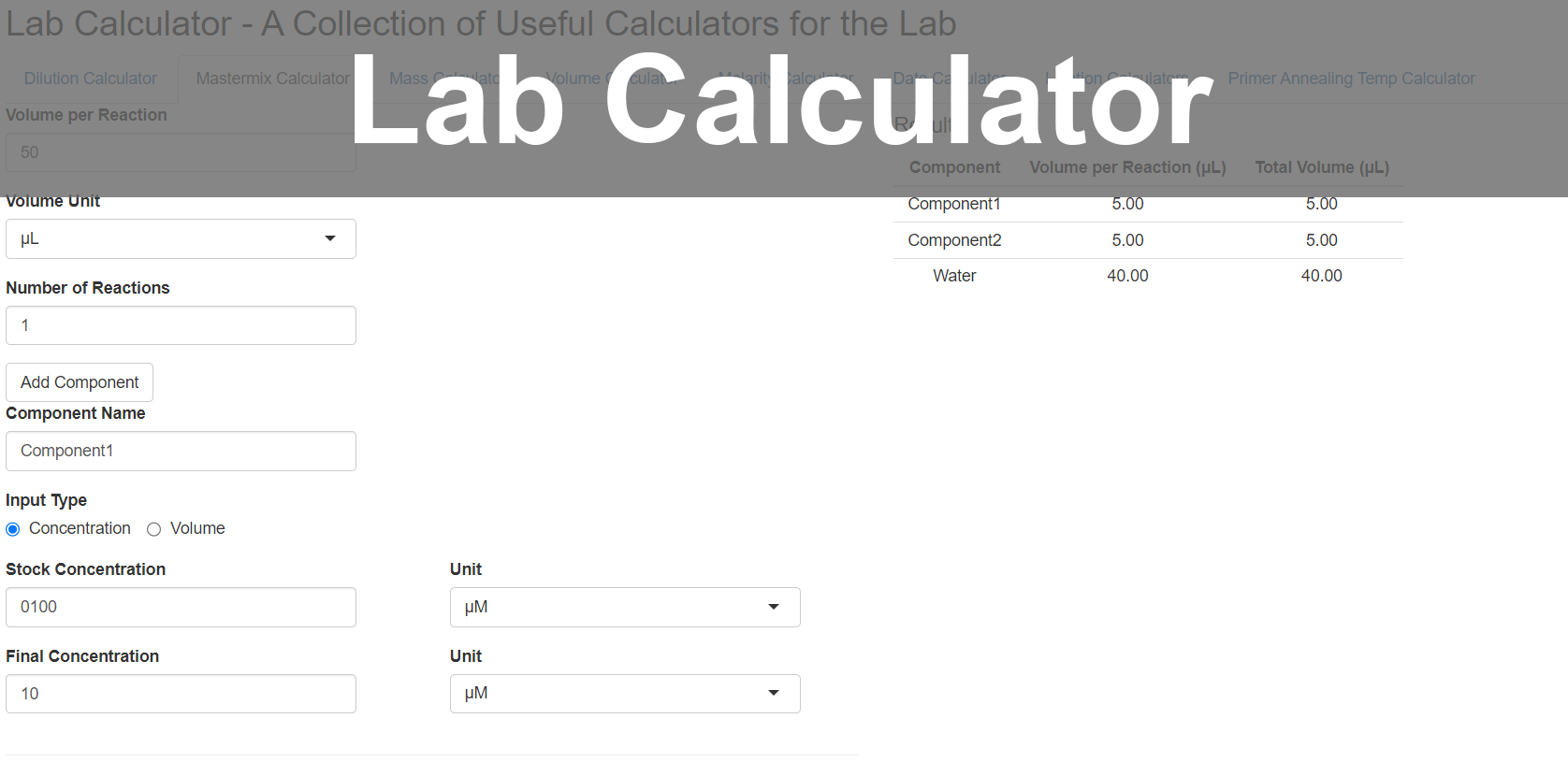
In my personal time, I've created multiple apps such as to analyze search trends in Google search history, or perform regular lab calculations, using 'shiny' in R.
As a Ph.D. holder in Cellular and Molecular Biology, I've worked with top-notch institutions like the University of Bern and Tata Institute of Fundamental Research. My publications in top-tier journals and oral presentations at international conferences reflect my enthusiasm to communicate.

Bioinformatics Scientist
Get to know me better! Here are some personal and professional details that define who I am.
- Birthday: 22nd September, 1989
- City: Bern, Switzerland
- Website: www.prateekarora.net
- Age: 35
- Degree: PhD
- Email: prateek@prateekarora.net
Facts
In addition to my expertise in experimental biology and bioinformatics, I have also achieved notable success in various areas, including publications, collaborative projects between multiple groups, web app development, and training and mentoring of students etc. The following section highlights some of my notable achievements:
Publications published or in preparation
Collaborative Projects within and outside the group
Web Apps developed and hosted
Students Trained for Masters/Bachelors dissertation
Skills
In the dynamic field of bioinformatics, I have honed a diverse skill set that enables me to tackle complex biological questions with precision and innovation. My expertise spans a range of technical proficiencies including advanced programming in R and Python, developing and deploying Shiny apps, and leveraging Docker for efficient, reproducible research. I am adept in cloud computing, utilizing platforms like AWS to handle large-scale data analysis. Additionally, I possess strong competencies in experimental design, pipeline development, and multi-omics data integration. These skills, combined with my dedication to continuous learning and collaboration, empower me to drive impactful research and deliver insightful, data-driven solutions.
Resume
Here you can find a brief overview of my professional journey and academic qualifications.
You can download my full resume in Pdf format here.
Summary
Prateek Arora
Passionate bioinformatician with expertise in single-cell transcriptomics, spatial omics, metabolomics, multi-omics, shiny apps, and pipeline development. Proven track record in mining, analysing and effectively visualizing available information to facilitate target discovery. Known for deep understanding of biology, attention to details, collaborative projects, and strong written and oral communication skills. With experience of handling multiple projects and teams in academic and industrial research, I look forward to new challenging roles.
Education
PhD (M.Sc. and PhD) - Cellular and Molecular Biology
2020
Tata Institute of Fundamental Research, Mumbai, India
My project involved, designing experiments, collecting data, analyzing data, performing statistical tests, regression fittings etc. By the project I established various analysis paradigms in the research group. This work was published in an international peer reviewed journal.
In past I have also worked on a project which involved study of role of Canonical Wnt signalling regulating laminin levels(Extracellular matrix molecule) in the development of zebrafish appendage -fin fold(limb homologue).
B.Sc. (H) Microbiology
2011
Ram Lal Anand College,
University of Delhi, New Delhi, India
Conferences and Publications
Oral Presentations
Shiny Conf 2024
Swiss Zebrafish Meeting 2023
Selected publications
Coppe, B., Galardi-Castilla, M., Sanz-Morejón, A., Arora, P.*, et al. Circulation, 2025- https://doi.org/10.1161/CIRCULATIONAHA.124.070323
Garcia-Poyatos, C., Arora, P.*, et al, Dev Cell, 2024- https://doi.org/10.1016/j.devcel.2024.04.012
Chavez, M. N., Arora, P.*, et al, iScience, 2024- https://10.1016/j.isci.2024.111406
Marques, I. J., Ernst, A.#, Arora, P.#*, et al, Development, 2022- https://doi.org/10.1242/dev.200375
Trainings and Certifications
Introduction to Machine Learning with Python (SIB- Swiss Institute of Bioinformatics)
Long-read sequence analysis (SIB- Swiss Institute of Bioinformatics)
Professional Experience
RESEARCH ASSOCIATE - BIOINFORMATICS
July 2023 - Present
University of Bern, Bern, Switzerland
- Managed 5 projects simultaenously while training 2 students
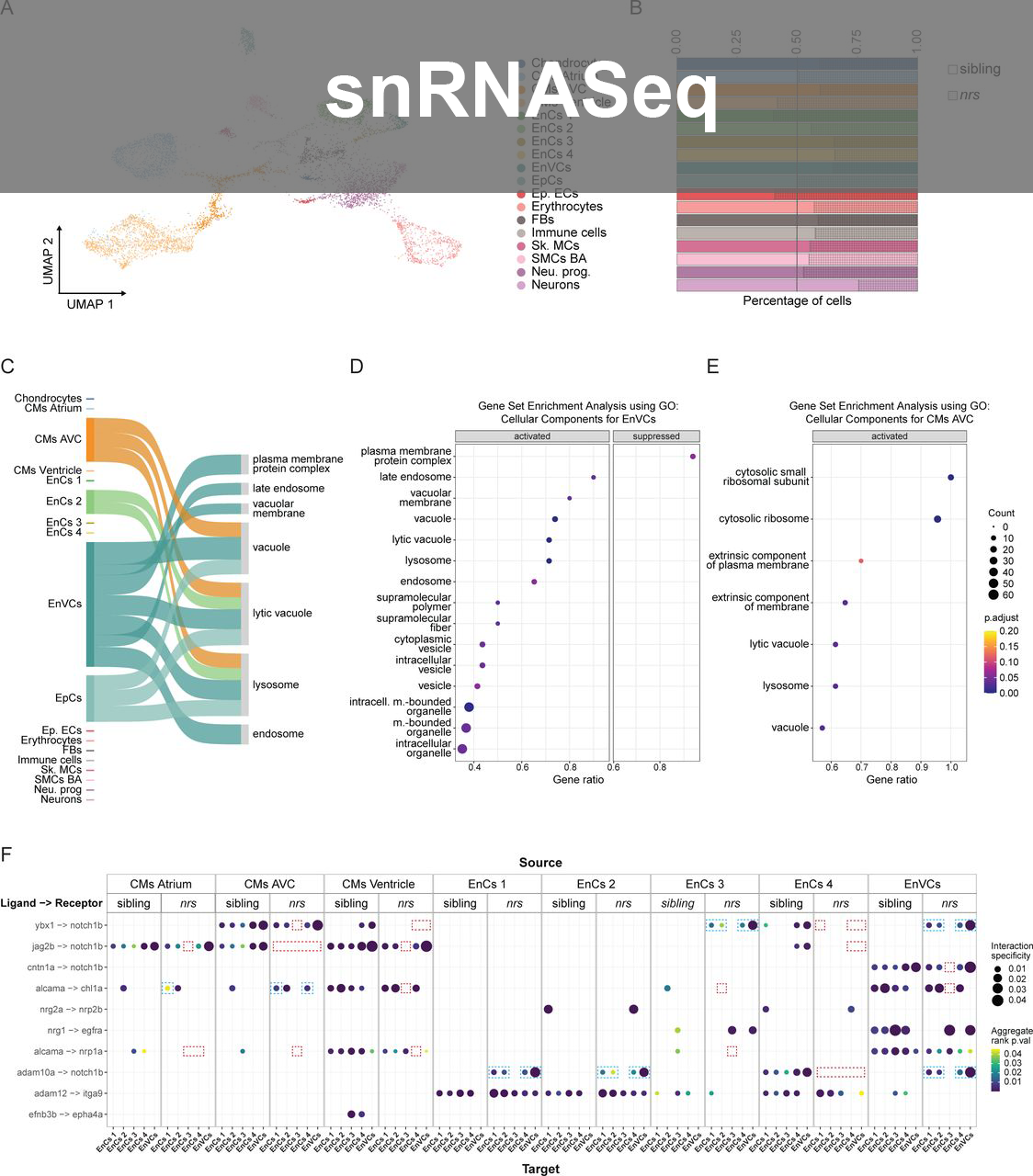
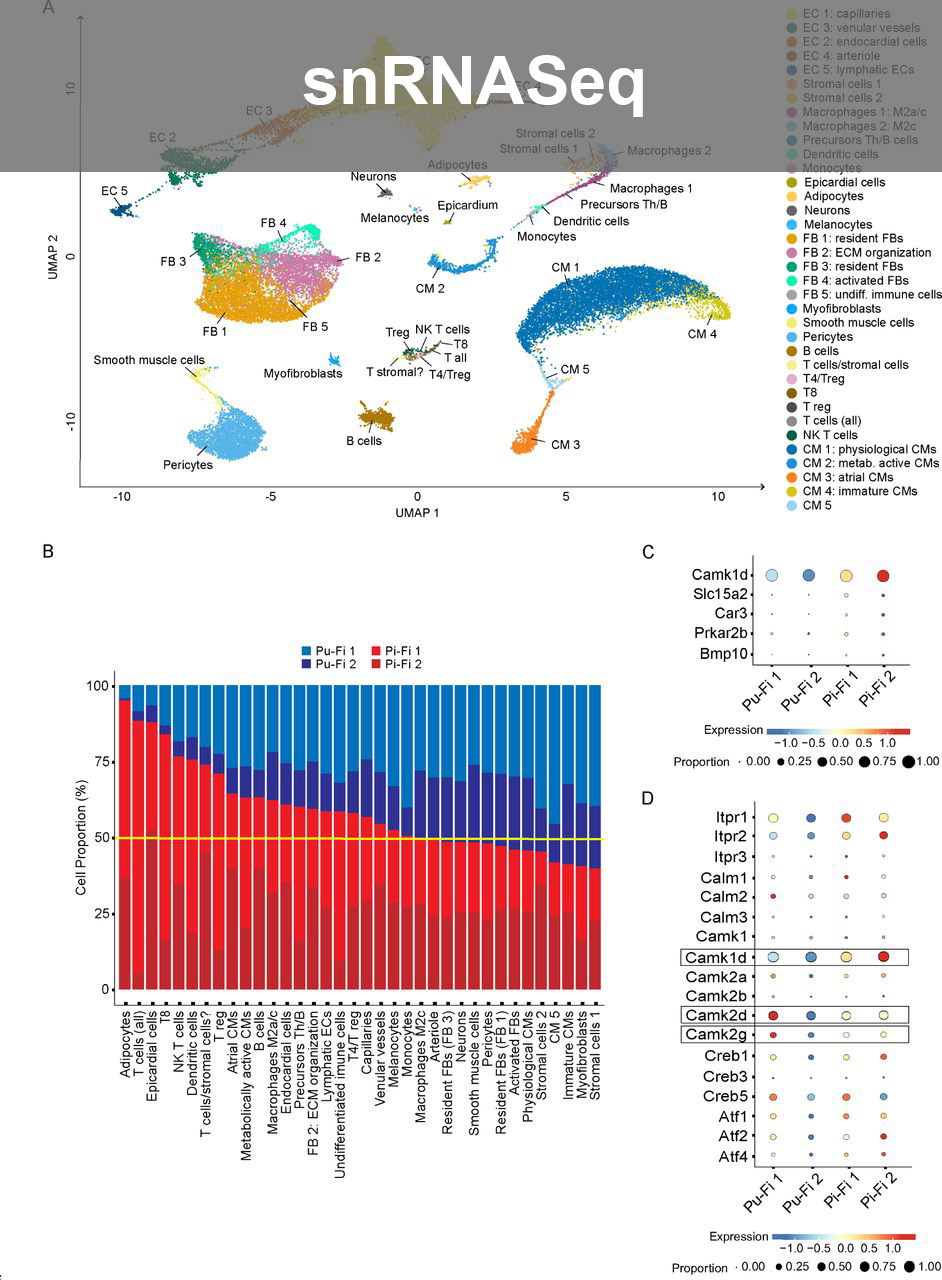
- Long-read snRNAseq Analysis: First group in Switzerland to perform Long-read snRNASeq using Oxford Nanopore Sequencing; designed experiments and uncovered novel splice variants in regenerating zebrafish hearts; analysis was performed on HPC cluster using Seurat, Pseudobulk differential analysis, LIANA, scDblFinder, OmniPath on self-maintained Docker containers. Supervised PhD student.
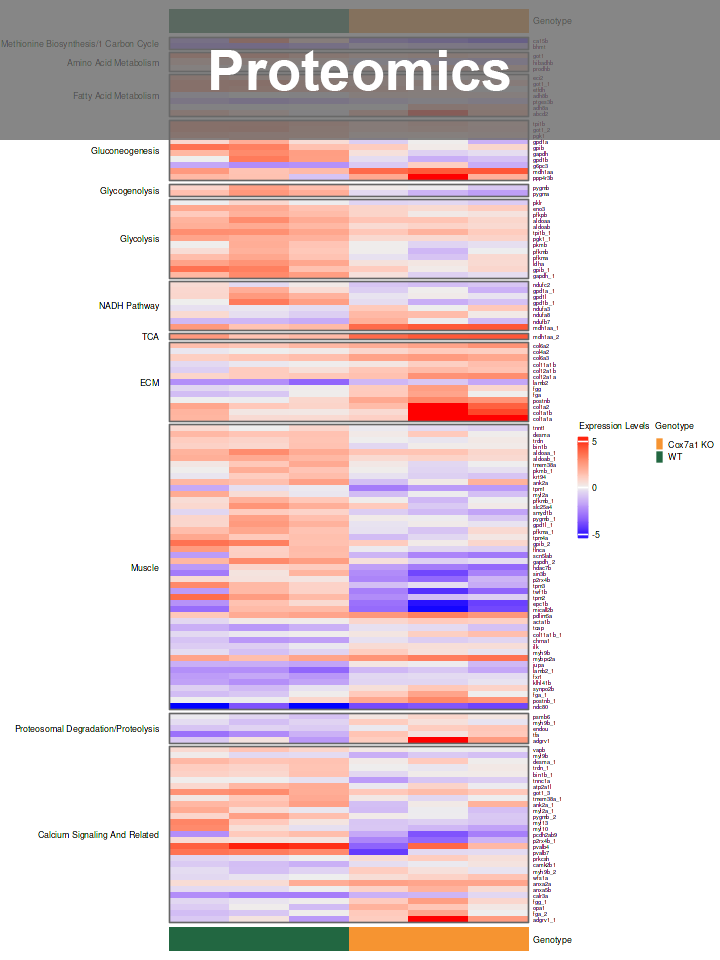
- Proteomics Analysis: Explored mitochondrial proteome in zebrafish muscle and heart mutants, utilizing Limma for differential analysis, GO, GSEA for pathway analysis and NLP for literature review.
- Spatial-transcriptomics: Showed evidence for in situ vaccination in Nanostring panels in mice to study high dose X-ray microbeams using Pathways analysis with SingleR, Clusterprofiler, ComplexHeatMaps; collaboration between 2 research groups
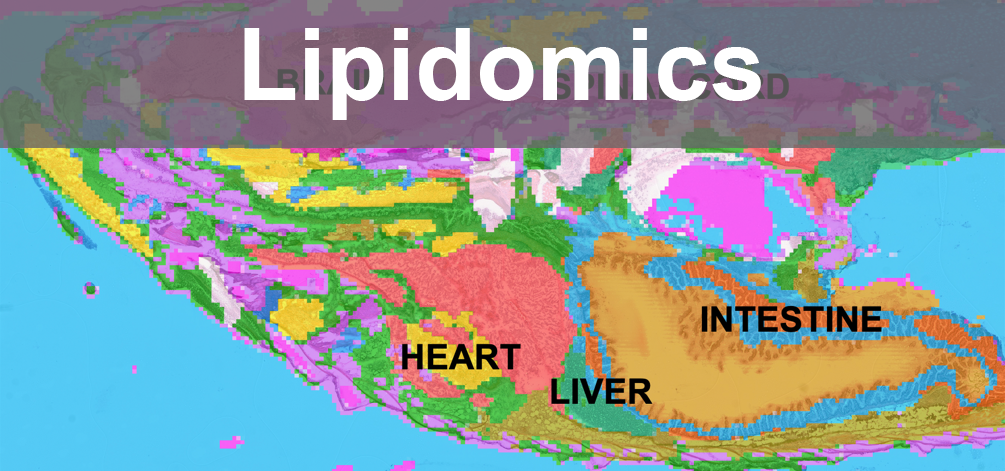
- Pipeline Development- Lipidomics: Innovated a pipeline for Spatial Mass Spectrometry Imaging Lipidomics in zebrafish, incorporating statistical tests, libraries as Cardinal and MetaboAnnotation which reduced processing time from 3 months to 3 days
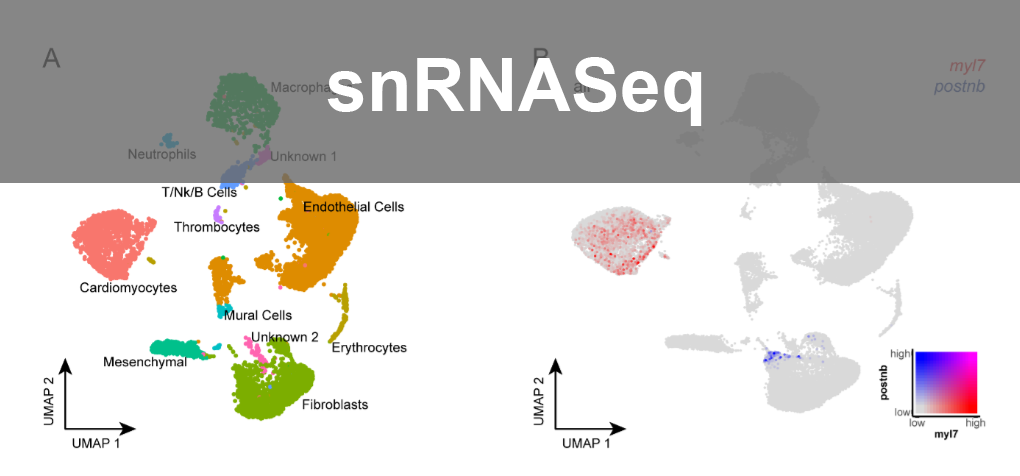
- Single-cell Analysis: Analysed developing mouse heart and mutants using 10X Genomics, presenting findings to collaborating groups and made user friendly shiny app.
- Methylation Analysis: Discovered methylation differences across generations in mouse after cardiac injury using Infinium Mouse Methylation BeadChip.
POSTDOCTORAL FELLOW - BIOINFORMATICS
January 2021 - June 2023
University of Bern, Bern, Switzerland
- snRNAseq Analysis: Advanced autophagy model studies in zebrafish hearts using Seurat and LIANA (ligand receptor); explored inheritance patterns in mouse hearts. Pseudobulk analysis using Libra, shiny app with ShinyCell, Pathway analysis.
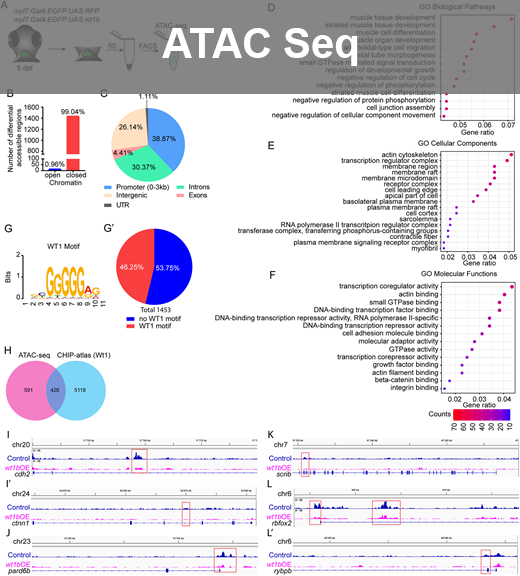
- ATACseq Studies: Investigated WT1 transcription factor's role in zebrafish and mouse sperm using pipeline tool - snakemake and various bioinformatics tools- Bowtie, picardtools, MACS3, Samtools, Genrich, Bedtools. Found critical transcription factors using CHIPSeeker, DiffBind, MEME suite for transcription factor site discovery etc.
- Target Discovery Pipeline: Developed a Ligand Receptor Network pipeline using bulk RNAseq, DESeq2, Network theory, leading to 2 PhD projects.
- Meta-analysis: Conducted comprehensive meta-analysis of Bulk RNAseq Data for regenerating hearts, uncovering conserved pathways and training PhD students.
SENIOR BIOINFORMATICIAN
June 2019 - June 2020
Elucidata Data Consulting Pvt Ltd, New Delhi, India
- Drug Target Discovery: Led a team to develop a drug target discovery pipeline, combining bulk and scRNAseq datasets on AWS; 20 potential targets identified.
- Team Leadership: Line manager of a team of two, task assignment, oversaw internal product development, validated scientific products, and set departmental OKRs.
Portfolio
Welcome to my portfolio! Here, you'll find a selection of my most impactful projects that demonstrate my expertise in bioinformatics and computational biology. From single-cell RNA sequencing in Switzerland to developing intuitive Shiny apps for data visualization, my work spans various innovative areas in the field. I have led advanced ATACseq studies to uncover key transcription factors, performed meta-analyses to reveal conserved pathways in heart regeneration, and created efficient pipelines for spatial lipidomics. Each project highlights my commitment to advancing scientific discovery through robust data analysis and collaborative efforts.
- All
- Single-cell/ Bulk RNASeq
- Proteomics/Lipidomics/ATACseq
- Other Shiny Apps
Contact
Get in touch! Whether you have questions, collaboration ideas, or just want to connect, feel free to reach out using the form below or through my social profiles.
Location
University of Bern, Switzerland